plot_pca_component_variance#
- scikitplot.api.decomposition.plot_pca_component_variance(clf, *, target_explained_variance=0.75, model_type=None, title='Cumulative Explained Variance Ratio by Principal Components', title_fontsize='large', text_fontsize='medium', x_tick_rotation=0, **kwargs)[source]#
Plots PCA components’ explained variance ratios. (new in v0.2.2)
Added in version 0.2.2.
- Parameters:
- clfobject
PCA instance that has the
explained_variance_ratio_attribute.- target_explained_variancefloat, optional, default=0.75
Looks for the minimum number of principal components that satisfies this value and emphasizes it on the plot.
- titlestr, optional, default=’Cumulative Explained Variance Ratio by Principal Components’
Title of the generated plot.
- title_fontsizestr or int, optional, default=’large’
Font size for the plot title. Use e.g., “small”, “medium”, “large” or integer-values.
- text_fontsizestr or int, optional, default=’medium’
Font size for the text in the plot. Use e.g., “small”, “medium”, “large” or integer-values.
- x_tick_rotationint, optional, default=0
Rotates x-axis tick labels by the specified angle.
Added in version 0.3.9.
- **kwargs: dict
Generic keyword arguments.
- Returns:
- axmatplotlib.axes.Axes
The axes on which the plot was drawn.
- Other Parameters:
- axmatplotlib.axes.Axes, optional, default=None
The axis to plot the figure on. If None is passed in the current axes will be used (or generated if required).
Added in version 0.4.0.
- figmatplotlib.pyplot.figure, optional, default: None
The figure to plot the Visualizer on. If None is passed in the current plot will be used (or generated if required).
Added in version 0.4.0.
- figsizetuple, optional, default=None
Width, height in inches. Tuple denoting figure size of the plot e.g. (12, 5)
Added in version 0.4.0.
- nrowsint, optional, default=1
Number of rows in the subplot grid.
Added in version 0.4.0.
- ncolsint, optional, default=1
Number of columns in the subplot grid.
Added in version 0.4.0.
- plot_stylestr, optional, default=None
Check available styles with “plt.style.available”. Examples include: [‘ggplot’, ‘seaborn’, ‘bmh’, ‘classic’, ‘dark_background’, ‘fivethirtyeight’, ‘grayscale’, ‘seaborn-bright’, ‘seaborn-colorblind’, ‘seaborn-dark’, ‘seaborn-dark-palette’, ‘tableau-colorblind10’, ‘fast’].
Added in version 0.4.0.
- show_figbool, default=True
Show the plot.
Added in version 0.4.0.
- save_figbool, default=False
Save the plot.
Added in version 0.4.0.
- save_fig_filenamestr, optional, default=’’
Specify the path and filetype to save the plot. If nothing specified, the plot will be saved as png inside
result_imagesunder to the current working directory. Defaults to plot image named to usedfunc.__name__.Added in version 0.4.0.
- overwritebool, optional, default=True
If False and a file exists, auto-increments the filename to avoid overwriting.
Added in version 0.4.0.
- add_timestampbool, optional, default=False
Whether to append a timestamp to the filename. Default is False.
Added in version 0.4.0.
- verbosebool, optional
If True, enables verbose output with informative messages during execution. Useful for debugging or understanding internal operations such as backend selection, font loading, and file saving status. If False, runs silently unless errors occur.
Default is False.
Added in version 0.4.0: The
verboseparameter was added to control logging and user feedback verbosity.
Examples
>>> from sklearn.decomposition import PCA >>> from sklearn.datasets import load_digits as data_10_classes >>> import scikitplot as skplt >>> X, y = data_10_classes(return_X_y=True, as_frame=False) >>> pca = PCA(random_state=0).fit(X) >>> skplt.decomposition.plot_pca_component_variance( ... pca, target_explained_variance=0.95 ... )
(
Source code,png)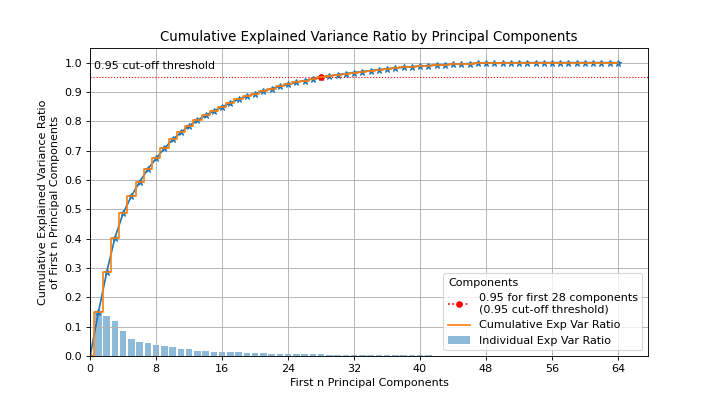
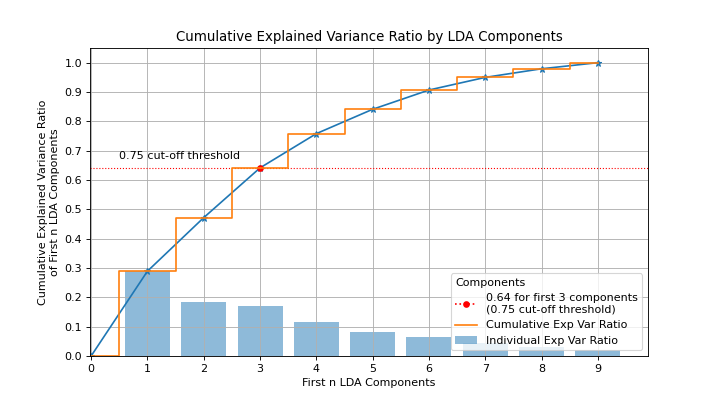
>>> from sklearn.discriminant_analysis import ( ... LinearDiscriminantAnalysis, ... ) >>> from sklearn.datasets import load_digits as data_10_classes >>> import scikitplot as skplt >>> X, y = data_10_classes(return_X_y=True, as_frame=False) >>> clf = LinearDiscriminantAnalysis().fit(X, y) >>> skplt.decomposition.plot_pca_component_variance(clf)
(
Source code,png)