plot_silhouette with examples#
An example showing the plot_silhouette function
used by a scikit-learn clusterer.
# Authors: The scikit-plots developers
# SPDX-License-Identifier: BSD-3-Clause
Import scikit-plots#
from sklearn.cluster import KMeans
from sklearn.datasets import (
load_iris as data_3_classes,
)
from sklearn.model_selection import train_test_split
import numpy as np
np.random.seed(0) # reproducibility
# importing pylab or pyplot
import matplotlib.pyplot as plt
# Import scikit-plot
import scikitplot as sp
Loading the dataset#
# Load the data
X, y = data_3_classes(return_X_y=True, as_frame=False)
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.5, random_state=0)
Model Training#
# Create an instance of the LogisticRegression
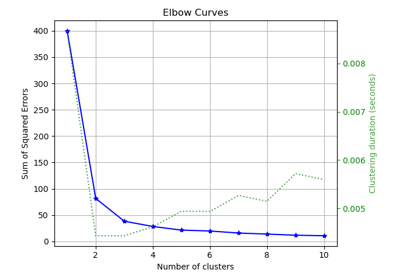
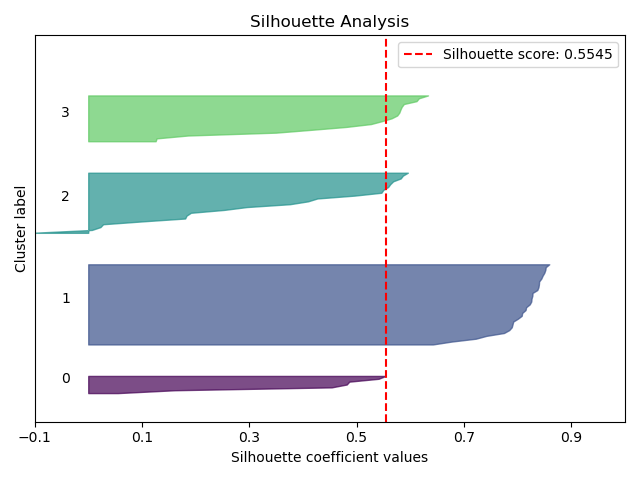
model = KMeans(n_clusters=4, random_state=0)
cluster_labels = model.fit_predict(X_train)
Plot!#
# Plot!
ax = sp.metrics.plot_silhouette(
X_train,
cluster_labels,
save_fig=True,
save_fig_filename="",
# overwrite=True,
add_timestamp=True,
# verbose=True,
)
Total running time of the script: (0 minutes 0.281 seconds)
Related examples